Ph. D. Joel Ricci-López
- Sr. Data scientist at AB InBev, Grupo Modelo.
- More than six years of experience working on data science and machine learning:
- Applied to retail sector, biological sciences, and molecular modeling.
- Experience on Python, R, SQL, Scikit-Learn, PySpark, TensorFlow, Azure Databricks, and PowerBI.
- Ph.D. in nanoscience and nanotechnology.
- Passionate about programming, artificial intelligence, web development, and graphic design.
- About me
- Portfolio
- Curriculum vitae
- Skills
About me
About me
Hi, I’m Joel.
I was born in Oaxaca, México 
I hold a Ph.D. on Nanoscience, and a Master’s and Bachelor’s degree in Life Sciences 
I have experience as a freelance web developer, data analyst, research assistant, and teacher.
During my career, I have developed skills in critical thinking, research, collaboration, problem-solving, project management, and self-organization.
I really love learning new things every day; programming, science, artificial intelligence, graphic design, etc.
I enjoy drawing, painting, and digital designing.
I love reading, especially science fiction and fantasy. Greg Egan, Carl Sagan, and George R. R. Martin are at the top of my favorite authors. I also like watching anime, playing chess, and playing video games.

My Projects
- All
- #MachineLearning
- #DataVisualization
- #DataScience
- #WebDevelopment
- #WebApp
- #Publication
ML+Ensemble Docking Repository
A GitHub repository containing scripts and notebooks to perform molecular modeling, data analysis, and machine learning modeling. Implemented in Ricci-Lopez, et al. (2021).
My Obsidian.md digital garden
My digital garden 🍀 is a personal wiki that contains notes (taken with Obsidian.md) related to machine learning and data science. Deployed on Netlify using Jekyll. GitHub
Interactive Scientific Poster
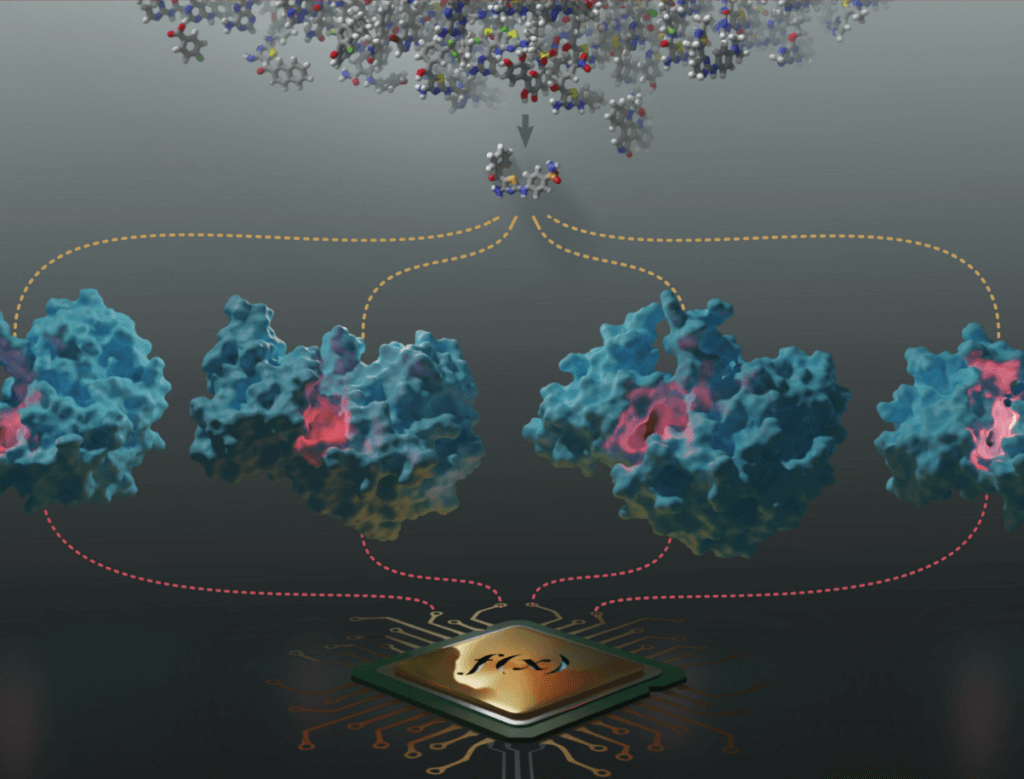
A web app with interactive molecular and data visualizations to show the results of a CNN model implemented for protein druggability prediction. Awarded first place during #LatinXChem2021. Developed using Python and Dash. GitHub
#Python #DataVisualization #WebApp #MolecularModeling #MachineLearning
Web page: "Molecular Modeling Workshop"
A web page with content from the “Molecular Modeling Workshop“, taught by me to postgraduate students (teaching assistant, Prof. Sergio Aguila). The page includes tutorials related to data analysis using Bash and Python. Developed using R, Markdown, and JavaScript.
Web page: "HPV: Molecular Virtual Screening"
A web page describing the methodology and results from my M.Sc. thesis and the scientific publication: Ricci-Lopez, et al. (2019). PlosOne.
#R #DataScience #DrugDiscovery #MolecularModeling #Bash #Python
ConvNets: Pneumonia detection from X-ray images
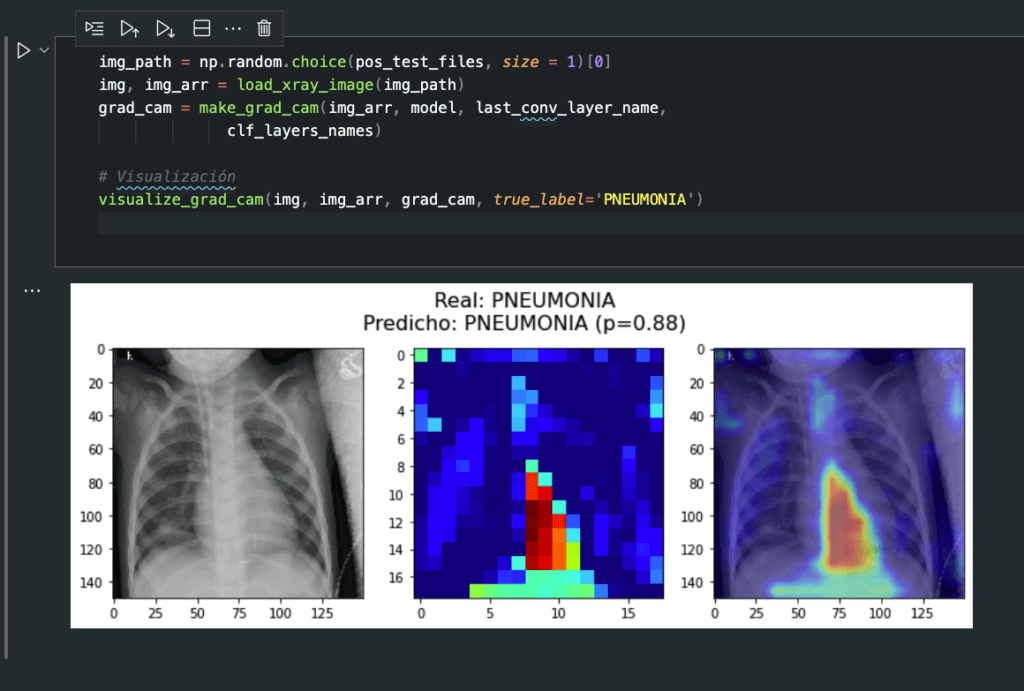
An implementation of Convolutional Neural Networks for Pneumonia prediction using X-ray images. Includes preprocessing steps, Keras implementation, Transfer Learning using the VGG16 network, and model explainability using the GradCAM method.
#MachineLearning #Python#DeepLearning#CNN #Keras #Tensorflow #TransferLearning
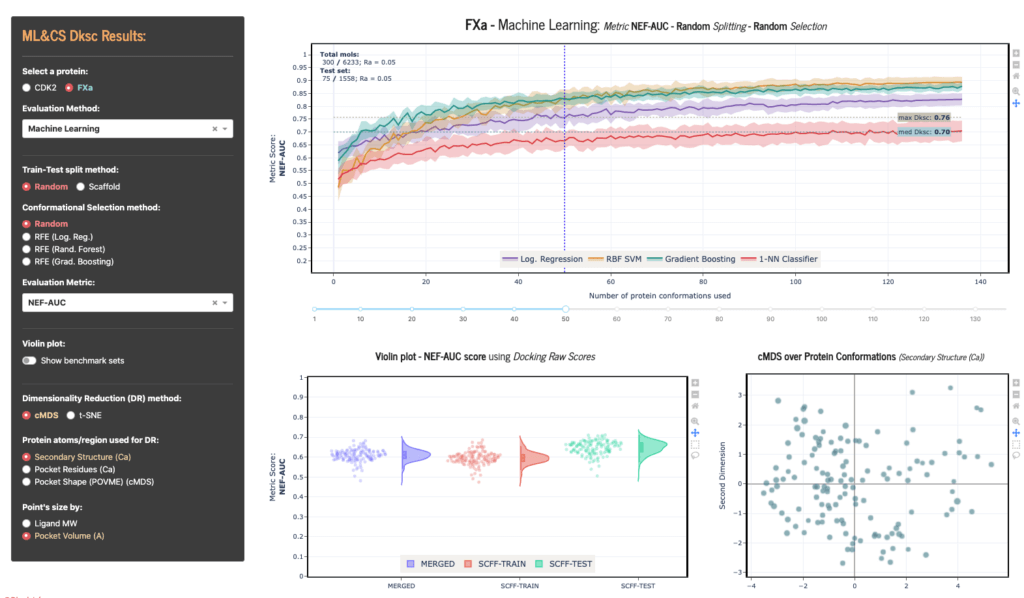
Dashboard: Analyzing Molecular Docking results
A web app with interactive plots designed to evaluate the results of integrating Machine Learning with Ensemble Docking simulations (in-house dataset). Developed using Python, Dash and Plotly.
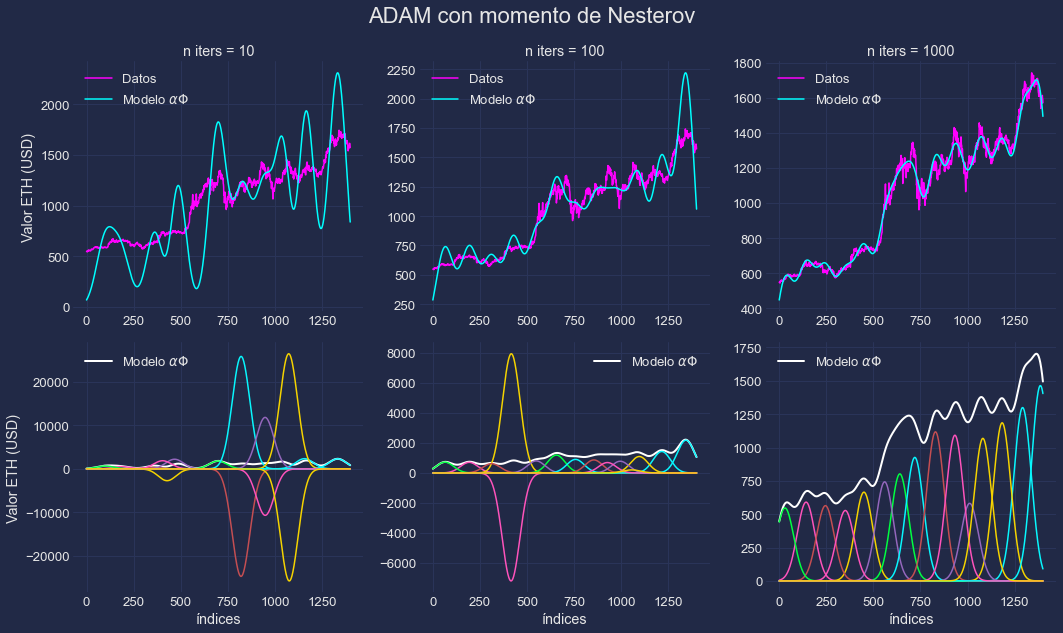
Gradient Descent Algorithms from Scratch
Implementation of Gradient Descent Algorithms from scratch using python: GD, Stochastic GD, Nesterov, AdaDelta, and ADAM.
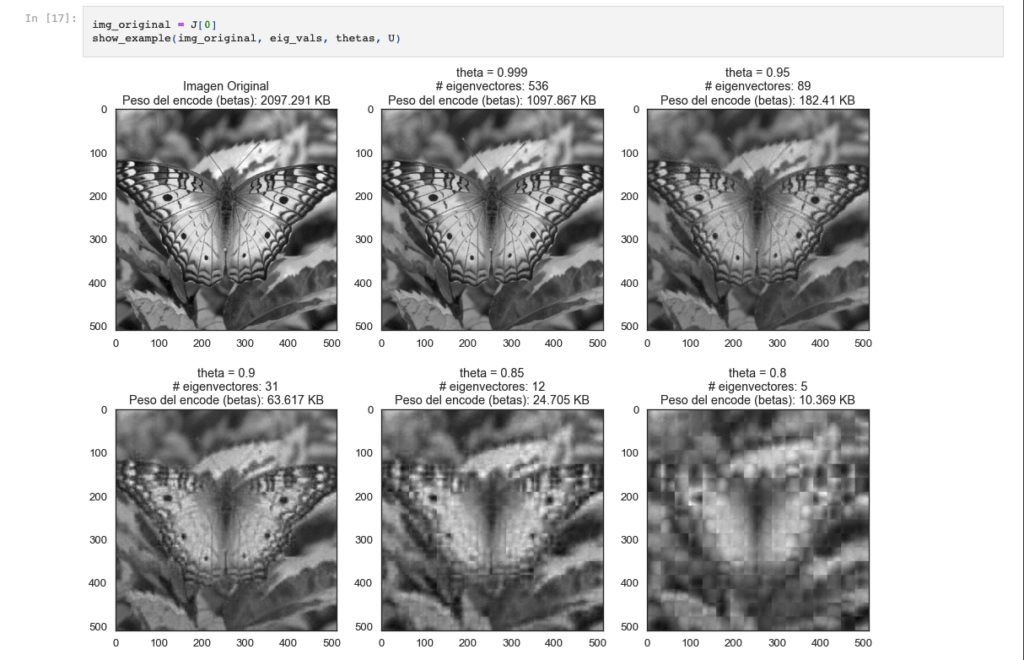
Image Compression using PCA
A colab notebook explaining how to perform image compression 🎑 using Principal Component Analysis.
#MachineLearning #Jupyter #Colab #Python
#NumPy #ScikitLearn
Web page: "LVMM"
A web page developed for the LVMM using WordPress.
Web page: "AguilaLab"
A web page developed for the AguilaLab Team. Developed using WordPress.
Current position:
- Senior Data Scientists.
- AB-InBev. July 2022 – present.
- Created Market Basket Analysis models using Python to feed Power BI interactive dashboards.
- Used Python and PowerBI to develop dashboards related to Customer Retention, RFM Analysis, and Cohort Analysis.
- Utilized Azure Databricks, PySpark, MLFlow, and Azure ML to predict Customer Churn using Machine Learning.
- Utilized SQL, Python, and Azure Databricks to perform exploratory data analysis to monitor and report sales-related KPIs.
- Using Azure Databricks to ingest, integrate, and validate data from multiple data sources.
- Designed and implemented automated text-based SKU/product matching pipelines using Azure Databricks, Python/PySpark and Azure ML to compare product prices among different marketplaces.
- AB-InBev. July 2022 – present.
Professional experience:
- Ph.D. Researcher in Nanoscience and Bioinformatics.
- Ensenada Center for Scientific Research and Higher Education (CICESE) and Nanoscience & Nanotechnology Centre (CNyN), UNAM. April 2018 – November 2023.
- Thesis title: “Virtual screening using machine learning techniques and ensemble docking-based molecular descriptors”.
- Ph.D. Advisors: Dr. Carlos A. Brizuela and Sergio A. Águila.
- Brief Synopsis of Research:
- We aimed to test and design a structure-based virtual screening pipeline combing molecular docking and machine learning to improve molecular virtual screening performance.
- We employ High-Performance Computing resources, parallel computing, data analysis, and machine learning models to improve computer-aided drug discovery.
- Ensenada Center for Scientific Research and Higher Education (CICESE) and Nanoscience & Nanotechnology Centre (CNyN), UNAM. April 2018 – November 2023.
- Visiting Research Scientist.
- HPC-Europa3 EU project. March – April 2022
- Participated in the EC-funded HPC-Europa3 Transnational Access Programme, collaborating with the High-Performance Computing facilities at EPCC, University of Edinburgh.
- Utilized the Cirrus supercomputing cluster at the University of Edinburgh for intensive parallel computing in molecular simulations, leveraging both CPUs and GPUs to accelerate computational tasks related to molecular dynamics simulations.
- Utilized Python data science libraries, Jupyter notebooks, visualization packages, and unsupervised machine learning algorithms to analyze molecular dynamics data.
- HPC-Europa3 EU project. March – April 2022
- Research Assistant – Data Analyst
- At Red nanoFAB. January – April 2018.:
Working on molecular modeling and simulation of bionanomaterials under the supervision of Dr. Sergio A. Águila (redesnanofab@gmail.com).- Worked on molecular modeling, simulation of bionanomaterials, and exploratory data analysis.
- Employed python and bash scripting to perform compute-intensive simulations, modeling and data analytics workflows related to molecular modeling data, generated using HPC resources.
- At Red nanoFAB. January – April 2018.:
- Teaching
- Assistant lecturer in Algorithms for Bioinformatics:
Lectures related to Molecular Biology and Protein Bioinformatics. Under the supervision of Dr. Carlos A. Brizuela Rodríguez (cbrizuela@cicese.mx). M.Sc. in Computer Science, CICESE.
January to February – 2022.
- Conducted sessions on data analysis using Python, R, and Bash to master’s level students.
- Assistant lecturer in Simulation of biological systems: Workshop on molecular dynamics and molecular docking simulations. Under the supervision of Dr. Sergio A. Águila. M.Sc. in Nanoscience and Nanotechnology, CNyN, UNAM. Workshop website . January to May – 2020 and 2021.
- Assistant lecturer in Bioinformatics: Lectures and workshops related to Protein Bioinformatics. Under the supervision of Dr. M. Asunción Lago (alago@cicese.mx). M.Sc. in Life Sciences, CICESE. January to February – 2019, 2020, and 2021.
- Assistant lecturer in Algorithms for Bioinformatics:
Lectures related to Molecular Biology and Protein Bioinformatics. Under the supervision of Dr. Carlos A. Brizuela Rodríguez (cbrizuela@cicese.mx). M.Sc. in Computer Science, CICESE.
January to February – 2022.
- Research Internship
- High-performance molecular dynamics simulations and computational chemical studies:
Under the supervision of Dr. Joel B. Alderete y la Dr. Verónica A. Jiménez Curihual (veronica.jimenez@unab.cl).
Department of Chemical Sciences of Universidad Andrés Bello, Chile.
March – April 2017.
- High-performance molecular dynamics simulations and computational chemical studies:
Education:
- Ph.D. Student in Nanoscience
- Center for Scientific Research and Higher Education at Ensenada, B. C. México.
Thesis: Virtual screening using machine learning techniques and ensemble docking-based molecular descriptors.
2018 – 2023
- Center for Scientific Research and Higher Education at Ensenada, B. C. México.
- Deep Learning Summer School
- Neuromatch Academy.
International student in the Neuromatch Academy’s Deep Learning interactive course.
120 hours of theoretical and practical projects on Neural Networks, CNNs, Sequential and Generative Models, and Reinforcement Learning.
August 2021
- Neuromatch Academy.
- Artificial Intelligence Product Manager, Nanodegree
- Udacity (view certificate )
Nanodegree focused on the implementation and evaluation of the business value of an AI product.
– Data acquisition and data curation using Appen.
– ML models implementation using Google Cloud and AutoML.
– AI project proposal focused on Drug Discovery.
2020
- Udacity (view certificate )
- MolSSI Software Summer School.
- Best practices in software engineering – version control, continuous integration, data management and programming paradigms. By “The Molecular Sciences Software Institute” (MolSSI) at Texas Advanced Computing Center (TACC), Texas, USA.
July 2019.
- Best practices in software engineering – version control, continuous integration, data management and programming paradigms. By “The Molecular Sciences Software Institute” (MolSSI) at Texas Advanced Computing Center (TACC), Texas, USA.
- Master of Science degree in Life Sciences
- Center for Scientific Research and Higher Education at Ensenada, B. C. México.
Thesis: In silico study of Human Papillomavirus E6 protein to identify antiviral drugs.
Web page available here .
2015 – 2017
- Center for Scientific Research and Higher Education at Ensenada, B. C. México.
- Bachelor of degree in Biology
- Universidad del Mar, Campus Puerto Escondido. Oaxaca, México.
General exam to obtain bachelor’s degree in biology (EGEL-BIO).
2009 – 2014
- Universidad del Mar, Campus Puerto Escondido. Oaxaca, México.
Scientific publications:
- Research Articles
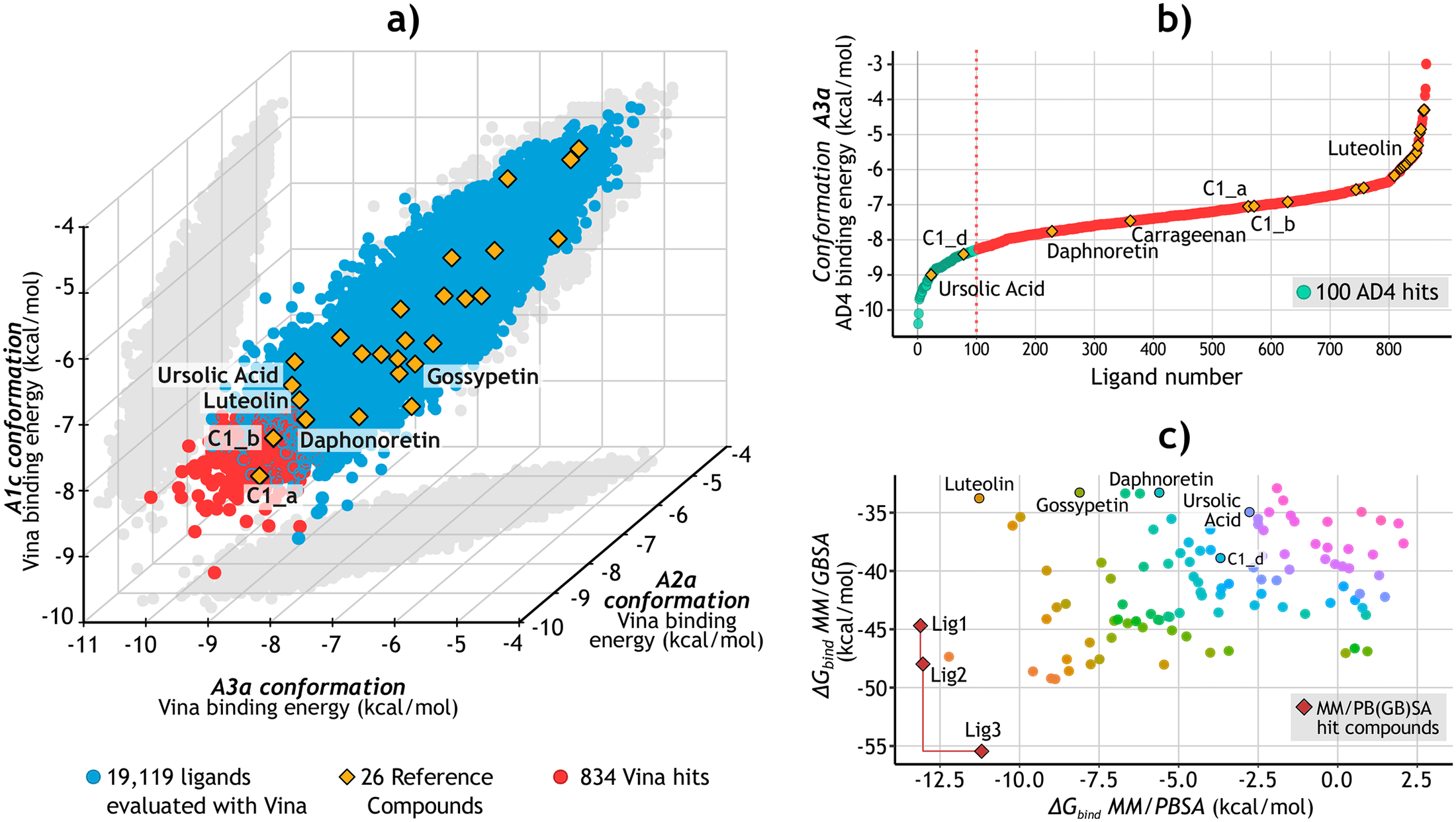
- Ricci-Lopez, J., Aguila, S. A., Gilson, M. K. & Brizuela, C. A. (2021). Improving Structure-Based Virtual Screening with Ensemble Docking and Machine Learning. J. Chem. Inf. Model., doi: 10.1021/acs.jcim.1c00511
- Ricci-Lopez, et al. (2019). Molecular modeling simulation studies reveal new potential inhibitors against HPV E6 protein. PLoS ONE 14(3), doi: 10.1371/journal.pone.0213028.
- Scientific Posters:
- Druggability prediction of protein conformations using convolutional neural networks.
At LatinXChem2021 Twitter Virtual Conference. Best poster award in the ChemBio category.
doi: 10.26226/morressier.616e5c2462ba8657678b132f
Interactive Poster.
September 2021. - Molecular modeling simulation studies reveal new potential inhibitors against HPV E6 protein. At Eighth Joint Sheffield Conference on Chemoinformatics, University of Sheffield, UK.
June 2019.
- Druggability prediction of protein conformations using convolutional neural networks.
- Oral presentations:
- Combining ensemble docking and machine learning to improve structure-based virtual screening.
At American Chemical Society Fall 2021: Resilience of Chemistry, Division of Chemical Information. Atlanta, Georgia, USA.
August 2021 - Refining structured-based virtual screening through ensemble docking and machine learning.
At III International Workshop On Computational Modeling of Biological Systems. Universidad del Quindio. Colombia.
November 2020. - Aprendizaje de máquina aplicado a los scores de acoplamiento molecular en conglomerado.
At III Coloquio de Simulaciones Computacionales en Ciencias, Universidad Nacional Autónoma de México, Ensenada, B. C., México. Best oral presentation award.
August 2020.
- Combining ensemble docking and machine learning to improve structure-based virtual screening.
Courses and Certificates:
- Courses and Workshops
- CABANA Workshop: Cheminformatics in Drug Discovery.
Five days workshop. Structure-based drug design and Ligand-based drug design.
By CABANA at CINVESTAV UGA LANGEBIO, México.
October 2019 - MOE workshop series: Ligand-Based Drug Design and SAR Analysis & Advanced Structure-Based Drug Design.
By Chemical Computing Group. California, USA.
September 2018 - Computational Biology oriented to drug design, III.
Two weeks course. Molecular docking and molecular dynamics simulations, fragment-based drug discovery, MD in mixed solvents and dynamic undocking, by CELFI Datos, University of Buenos Aires, Argentina.
May 2018
- CABANA Workshop: Cheminformatics in Drug Discovery.
- Online Courses
- Deep Learning Specialization (five courses).
By Coursera – DeepLearning.Ai. Certificate RK42DWNPUBD3
January 2022 - Applied Machine Learning with Python.
By Coursera – University of Michigan. Credential Y8494V5ZL6Y5
December 2021 - AWS Machine Learning.
By Udacity – AWS. Credential DG9NMPUA
October 2021 - Machine Learning.
By Coursera – Stanford. License 5C9XH7E8RDDT
June 2021 - Machine Learning Scientist with Python (96 hours).
By DataCamp. Credential 0236e8d560afd98ff6afbae32606b1c4f0369426
March 2021 - Using databases with Python.
By Coursera – University of Michigan. Credential NEJJRT599V8J
June 2019 - Introduction to Data Science in Python.
By Coursera – University of Michigan. Credential 6Z33HDXP553K
January 2018 - Python for Genomic Data Science.
By Coursera: Johns Hopkins University. Credential KT844NQQULF2
May 2016
- Deep Learning Specialization (five courses).
Technical Skills
Programming Languages:
PythonRDAXBashM (Power Query)JavaScript
Machine Learning:
Scikit-Learn- MLib PySpark
KerasTensorFlowMLflow
Data Science:
SQLPySparkDatabricksPandasNumPyTidyverseKNIMEAirflow
Data Visualization:
PowerBIDash (Python)Shiny (R)PlotlyMatplotlibBokehggplot2
Version Control and Development:
GitGitHubDocker
Web Development:
WordPressElementorHTML5CSS3React
Molecular Modelling:
ModellerNAMDAmberMDAutoDockVMDUCSF QuimeraBio3DRDKit
Graphic Design:
InkscapeBlenderGimp
Soft Skills
CommunicationCollaborationCritical thinkingCuriosityCreativityProject managementOrganizationStory Telling